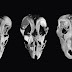
Some post-translational modifications of actin are present in all isoforms (structural variants) of the protein, whereas others are more specific.
Would you mind for a second
Monday, 14 January 2019
Protein modification fine-tunes the cell’s force producers
Some post-translational modifications of actin are present in all isoforms (structural variants) of the protein, whereas others are more specific.
Thursday, 23 June 2016
Precise control of brain circuit alters mood

By combining super-fine electrodes and tiny amounts of a very specific drug, researchers have singled out a circuit in mouse brains and taken control of it to dial an animal's mood up and down. (© mgkuijpers / Fotolia)
By combining super-fine electrodes and tiny amounts of a very specific drug, Duke University researchers have singled out a circuit in mouse brains and taken control of it to dial an animal's mood up and down.
Stress-susceptible animals that behaved as if they were depressed or anxious were restored to relatively normal behavior by tweaking the system, according to a study appearing in the July 20 issue of Neuron.
"If you 'turn the volume up' on animals that hadn't experienced stress, they start normal and then they have a problem," said lead researcher Kafui Dzirasa, an assistant professor of psychiatry and behavioral sciences, and neurobiology. "But in the animals that had experienced stress and didn't do well with it, you had to turn their volume up to get them back to normal. It looked like stress had turned the volume down."
The circuit the team identified and altered is a connection the prefrontal cortex uses to keep time for the limbic system, which governs emotions and basic drives. To regulate mood, the prefrontal cortex acts as a pacemaker to coordinate the actions of the amygdala, which governs stress responses, and the ventral tegmental area, which plays a role in the brain's reward circuitry.
"These subcortical circuits are the key regulators of our emotional life," said Helen Mayberg, a professor of psychiatry, neurology and radiology at Emory University who was not involved in this research. "What's great about this paper is that they use different approaches to see a circuit that's relevant to a lot of disorders," said Mayberg, who has been pioneering deep-brain stimulation of very specific sites in the human prefrontal cortex to treat mood disorders.
The emerging picture from this study and others is of a brain built of multi-part circuits that respond in concert and regulate one another. Specificity in understanding these circuits is going to be key to resolving different disorders, Dzirasa said.
"The prefrontal cortex is not just a blob of cells," Mayberg said. "These findings give insight into which cells go to which area and allow researchers to kind of choreograph their actions."
Dzirasa is an M.D. just finishing his residency in psychiatry and a Ph.D. neuroscientist with an engineering background. Postdoctoral researcher and first author Rainbo Hultman is a biochemist.
In addition to overcoming the challenges of understanding each other, they asked, "Could we go from a protein, to a signaling activity, to a cell, to a circuit, to this big activity that happens across the whole brain, to actual behavior?" Hultman said.
"Illness can happen at any one of these levels," said Dzirasa, who is also a member of the Duke Institute for Brain Sciences.
The team started by precisely placing arrays of 32 electrodes in four brain areas of the mice. Then they recorded brain activity as these mice were subjected to a stressful situation called chronic social defeat. This allowed them to see activity between the prefrontal cortex and three areas of the limbic system that are implicated in major depression.
To interpret the complicated data coming from the electrodes, the neuroscientists then turned to Duke colleagues David Dunson of statistical science and Lawrence Carin of electrical engineering, who specialize in statistical analysis of noisy data to find important patterns. Using machine learning algorithms, they identified which parts of the data seemed to be the timing control signal between the prefrontal cortex and the amygdala and zeroed in on the individual neurons involved in that circuit.
"They came back with, 'It's this clock signature here that is responsible for which mice become susceptible to stress and which become resilient,'" Dzirasa said.
Hultman then turned to engineered molecules called DREADD developed by University of North Carolina at Chapel Hill pharmacologist Bryan Roth. These Designer Receptors Exclusively Activated by Designer Drug are very specific signal receptors that can be incorporated into the neural circuit's control spots in very tiny amounts (0.5 microliter). A drug that attaches only to that DREADD is then administered to give the researchers control over the circuit.
This new combination of electronics and drugs to intervene in an individual brain circuit might be used to create mouse models of other mood disorders for other studies, Dzirasa said. But Emory's Mayberg cautions that a mouse brain is not a human brain and to assess anything like "mood" in a mouse, one can only infer from its behaviors. "It's hard to do, even in a human," she said.
Story Source:
The above story is based on materials provided by Duke University. Note: Materials may be edited for content and length.
Journal Reference:
Rainbo Hultman, Stephen D. Mague, Qiang Li, Brittany M. Katz, Nadine Michel, Lizhen Lin, Joyce Wang, Lisa K. David, Cameron Blount, Rithi Chandy, David Carlson, Kyle Ulrich, Lawrence Carin, David Dunson, Sunil Kumar, Karl Deisseroth, Scott D. Moore, and Kafui Dzirasa. Dysregulation of Prefrontal Cortex-Mediated Slow-Evolving Limbic Dynamics Drives Stress-Induced Emotional Pathology. Neuron, June 2016 DOI: 10.1016/j.neuron.2016.05.038
New study provides unprecedented insight into the fine details of neuronal communication
There are three main functions of the nervous system: sensory function, which detects changes in the body; integrative function, which makes decisions based on information it receives; and motor function, which carries electrical impulses to stimulate a response. In particular, the integrative functions of the brain bring sensory information together, add to memory, produce thoughts, and make decision.
The cerebellum, part of the brain that is responsible for motor control, serves as an optimal model system to study the integrative features of the nervous system at both the cellular and network level. The circuitry of the cerebellum is strikingly simple when compared with other regions of the brain, with a regularly repeating cellular organization across its outer layer. In addition, cerebellar-dependent tasks have been well-mapped to particular anatomical sub regions. Therefore, the cerebellum offers a unique opportunity to study the dynamics of how information is transferred and transformed within and between neurons to control motor behavior.
Neurons are divided in three main parts: dendrites, axons, and the soma. While dendrites receive and integrate synaptic input, axons transmit the resultant, compiled synaptic information to specific sites in the form of action potentials. Even within these structures there is sub-specialization; dendrites often support functional domains, or dendritic spines, multiplying their computational capacity. And axons achieve a high-degree of specificity in the organization and functional influence of sodium and potassium channels, despite the simplistic classical view of action potentials as monotypic binary pulses transmitted throughout the entire extent of an axon.
In their June 2016 publication in Neuron, researchers Matthew J.M. Rowan, Ph.D., and Jason M. Christie, Ph.D., describe how they overcame a major technical challenge precluding direct examination of axonal excitability. Because of its small diameter -- less than 500 nanometers -- the typical axon can't be examined by the conventional electrophysiological recordings. However, using optically-guided subcellular patching, in combination with organic actuators of neural activity, the scientists were able to sample targeted sub regions of axonal membrane including both presynaptic boutons and their attached axon shafts. This recording configuration allows for the direct assessment of the distribution and biophysical properties of ion channels and receptors expressed along an axon. And, in conjunction, it allows for the direct recording of neural signals including action potentials and subthreshold synaptic activity.
Notably, this study demonstrated that action potentials, often viewed as invariant pulses, are instead quite dynamic with their shape varying with subcellular location. The varicose geometry of boutons, alone, does not impose striking differences in spike duration. Rather, this physiology depends on the differential influence of potassium channel subtypes as well as a clustering of fast-activating potassium channels at presynaptic locations. The organizational feature described by this study allows axons to multiply their adaptive properties by tuning excitation in one axonal domain independent of other domains on an exquisitely local spatial scale, including between neighboring sites of release.
Future directions
According to Dr. Rowan, the clustered arrangement and variable expression density of potassium channels at boutons are key determinants underlying compartmentalized control of action potential width in a near synapse-by-synapse manner. Such organization yields a powerful adaptive property allowing individual release sites to locally inform the duration of a propagating spike, dependent on the local abundance of channels, and separate of other sites. Dr. Christie's research team will further investigate how spike signaling within axons is organized and modified, and the computational value of this organization to cerebellar microcircuits.
Story Source:
The above story is based on materials provided by Max Planck Florida Institute for Neuroscience. Note: Materials may be edited for content and length.
Journal Reference:
Matthew J.M. Rowan, Gina DelCanto, Jianqing J. Yu, Naomi Kamasawa, and Jason M. Christie. Synapse-level determination of action potential duration by K channels in axons.Neuron, June 2016 DOI: 10.1016/j.neuron.2016.05.035
Source : Science Daily
Friday, 17 June 2016
Mechanism for Rift Valley fever virus infection discovered

Brooke Harmon, a virologist at Sandia National Laboratories, led research that found a cellular pathway for Rift Valley fever virus infection, the first step in developing treatment for the highly infectious deadly disease. (Randy Wong, Sandia National Laboratories)
Viruses can't live without us -- literally. As obligate parasites, viruses need a host cell to survive and grow. Scientists are exploiting this characteristic by developing therapeutics that close off pathways necessary for viral infection, essentially stopping pathogens in their tracks.
Rift Valley fever virus (RVFV) and other members of the bunyavirus family may soon be added to the list of viruses denied access to a human host. Sandia National Laboratories researchers have discovered a mechanism by which RVFV hijacks the host machinery to cause infection. This mechanism offers a new approach toward developing countermeasures against this deadly virus, which in severe human infections causes fatal hepatitis with hemorrhagic fever, encephalitis and retinal vasculitis.
The results are reported in a paper, "A Genome-Wide RNAi Screen Identifies a Role for Wnt/Beta-Catenin Signaling During Rift Valley Fever Virus Infection," recently published in the Journal of Virology. The work was funded by Sandia's Laboratory Directed Research and Development program.
RVFV uses a cancer pathway
Little is known about the fundamental infection mechanisms and interactions between bunyaviruses and their host cells. Led by Sandia virologist Brooke Harmon, the researchers discovered that Wnt signaling is essential for bunyavirus infection.
The Wnt signaling pathway, which regulates critical cell processes, such as proliferation and differentiation, is already under heavy investigation by medical researchers because of its association with breast, melanoma, prostrate, lung, ovarian and other cancers and with Type II diabetes. Clinical trials are underway for cancer treatments targeting the Wnt pathway.
"We can take advantage of the work on cancer therapeutics. Inhibitors of this pathway are already being developed for several cancers. As those therapies move through clinical trials, we can apply them to infectious diseases," said Harmon.
Rift Valley a priority pathogen
You may not have heard of RVFV, but it's a familiar threat to anyone working in infectious diseases. The National Institute of Allergy and Infectious Diseases lists RVFV as a category A priority pathogen, meaning it poses the highest risk to national security and public health.
"Rift Valley combines some of the most sinister aspects of both Ebola and Zika into one virus," explained Harmon. "Like Ebola, it can cause hemorrhagic fever and be lethal within days of infection. Like Zika, it's transmitted by mosquitoes, can cause neurological disease in humans, and results in frequent miscarriages and fetal deformities in livestock."
Today RVFV predominantly affects animals, livestock in particular. Like most viruses transmitted by mosquitos, RVFV circulates predominantly in wild animals but has the potential to spill over into human populations, similar to avian influenza and West Nile virus.
While endemic to Africa, RVFV has spread to the Arabian Peninsula and has the capacity to emerge into further territories. Since the late 1990s, large-scale RVFV outbreaks in eastern and southern Africa, Mauritania, Saudi Arabia and Yemen have severely affected the health and economy of tens of thousands of humans and infected hundreds of thousands of livestock.
Bunyavirus family uses Wnt
Harmon and Sandia researcher Oscar Negrete began the project about five years ago by using high-throughput RNA interference to screen the entire human genome against RVFV. The researchers looked for genes that were required for virus infection, meaning that the virus cannot infect cells missing that gene.
From that initial screen, conducted at the University of California, Berkeley, and further screening at Sandia, they narrowed the field down to 381 genes of interest. "When we functionally clustered those genes, we found that the Wnt pathway was the most represented," said Negrete.
To test their hypothesis that the Wnt pathway is critical to RVFV infection, the researchers tested a vaccine strain of the virus. When those results supported their theory, they conducted the same experiments on wild type virus in a Biosafety Level-3 laboratory at Lawrence Livermore National Laboratory.
They expanded the testing to other members of the bunyavirus family like La Crosse virus and California encephalitis virus and found the same results. "This was somewhat unexpected because divergent bunyaviruses typically have their own unique features of infection. The fact that they shared this same pathway is exciting because it indicates Wnt signaling may be necessary to the virus family as a whole," said Negrete.
Getting ahead of the next big outbreak
The next step, said Negrete, is to further investigate the mechanisms of infection. The researchers also plan to look for other mechanisms of RVFV infection using CRISPR, or clustered regularly interspaced short palindromic repeats, which is complementary to RNA interference. This understanding can aid in the design of effective host-directed anti-viral therapeutics.
"We keep chasing these viruses. An outbreak like Zika happens and that's when the push begins for a therapeutic. We need to get out in front of the next big one because recent history has taught us that deadly diseases can rapidly spread from animals to humans and beyond endemic zones," said Harmon. "If there is an outbreak of RVFV or another bunyavirus, we hope to already have something in the arsenal."
Sandia National Laboratories is a multi-program laboratory operated by Sandia Corporation, a wholly owned subsidiary of Lockheed Martin Corp., for the U.S. Department of Energy's National Nuclear Security Administration. With main facilities in Albuquerque, N.M., and Livermore, Calif., Sandia has major R&D responsibilities in national security, energy and environmental technologies and economic competitiveness.
Story Source:
The above story is based on materials provided by DOE/Sandia National Laboratories. Note: Materials may be edited for content and length.
Journal Reference:
Brooke Harmon, Sara W. Bird, Benjamin R. Schudel, Anson V. Hatch, Amy Rasley, Oscar A. Negrete. A Genome-Wide RNAi Screen Identifies a Role for Wnt/Beta-Catenin Signaling During Rift Valley Fever Virus Infection. Journal of Virology, 2016; JVI.00543-16 DOI:10.1128/JVI.00543-16
Natural molecule could improve Parkinson's

These are brain scans of a representative patient showing Dopamine transporter binding (red) before and after 3-month NAC treatment. (Thomas Jefferson University)
The natural molecule, n-acetylcysteine (NAC), with strong antioxidant effects, shows potential benefit as part of the management for patients with Parkinson's disease, according to a study published in the journal PLOS ONE. Combining clinical evaluations of a patient's mental and physical abilities with brain imaging studies that tracked the levels of dopamine, the lack of which is thought to cause Parkinson's, doctors from the Departments of Integrative Medicine, Neurology, and Radiology, at Thomas Jefferson University showed that patients receiving NAC improved on both measures.
Current treatments for Parkinson's disease are generally limited to temporarily replacing dopamine in the brain as well as some medications designed to slow the progression of the disease process. Recently, researchers have shown that oxidative stress in the brain may play a critical role in the Parkinson's disease process, and that this stress also lowers levels of glutathione, a chemical produced by the brain to counteract oxidative stress. Studies in brain cells showed that NAC helps reduce oxidative damage to neurons by helping restore the levels of the antioxidant glutathione. NAC is an oral supplement that can be obtained at most nutrition stores, and interestingly also comes in an intravenous form which is used to protect the liver in acetaminophen overdose.
"This study reveals a potentially new avenue for managing Parkinson's patients and shows that n-acetylcysteine may have a unique physiological effect that alters the disease process and enables dopamine neurons to recover some function," said senior author on the paper Daniel Monti, M.D., M.B.A., Director of the Myrna Brind Center of Integrative Medicine, and the Brind-Marcus Center of Integrative Medicine at Thomas Jefferson University.
In this study, Parkinson's patients who continued their current standard of care treatment, were placed into two groups. The first group received a combination of oral and intravenous (IV) NAC for three months. These patients received 50mg/kg NAC intravenously once per week and 600mg NAC orally 2x per day on the non IV days. The second group, the control patients, received only their standard of care for Parkinson's treatment. Patients were evaluated initially, before starting the NAC and then after three months of receiving the NAC while the control patients were simply evaluated initially and three months later. The evaluation consisted of standard clinical measures such as the Unified Parkinson's Disease Rating Scale (UPDRS), a survey administered by doctors to help determine the stage of disease, and a brain scan via DaTscan SPECT imaging, which measures the amount of dopamine transporter in the basal ganglia, the area most affected by the Parkinson's disease process. Compared to controls, the patients receiving NAC had improvements of 4-9 percent in dopamine transporter binding and also had improvements in their UPDRS score of about 13 percent.
"We have not previously seen an intervention for Parkinson's disease have this kind of effect on the brain," said first author and neuro-imaging expert Andrew Newberg, M.D., Professor at the Sidney Kimmel Medical College at Jefferson and Director of Research at the Myrna Brind Center of Integrative Medicine. The investigators hope that this research will open up new avenues of treatment for Parkinson's disease patients.
Story Source:
The above story is based on materials provided by Thomas Jefferson University. Note: Materials may be edited for content and length.
Journal Reference:
Daniel A. Monti, George Zabrecky, Daniel Kremens, Tsao-Wei Liang, Nancy A. Wintering, Jingli Cai, Xiatao Wei, Anthony J. Bazzan, Li Zhong, Brendan Bowen, Charles M. Intenzo, Lorraine Iacovitti, Andrew B. Newberg. N-Acetyl Cysteine May Support Dopamine Neurons in Parkinson's Disease: Preliminary Clinical and Cell Line Data.PLOS ONE, 2016; 11 (6): e0157602 DOI:10.1371/journal.pone.0157602
Monday, 11 April 2016
New Microbes expands tree of life
Bacteria make up nearly two-thirds of all biodiversity on Earth, half of them uncultivable

- Laura A. Hug, Brett J. Baker, Karthik Anantharaman, Christopher T. Brown, Alexander J. Probst, Cindy J. Castelle, Cristina N. Butterfield, Alex W. Hernsdorf, Yuki Amano, Kotaro Ise, Yohey Suzuki, Natasha Dudek, David A. Relman, Kari M. Finstad, Ronald Amundson, Brian C. Thomas, Jillian F. Banfield. A new view of the tree of life. Nature Microbiology, 2016; 16048 DOI: 10.1038/nmicrobiol.2016.48
- Date:
- April 11, 2016
- Source:
- University of California - Berkeley
Sunday, 27 March 2016
Cells in standby mode

Normally, yeast cells are rod-shaped (above). After having their cell wall removed, however, they become spherical (below left). During dormancy (above right) the loss of the cell wall the cells retain their stability and keep their rod-shaped appearance (below right). (MPI f. Molecular Cell Biology and Genetics)
Normally, cells are highly active and dynamic: in their liquid interior, called the cytoplasm, countless metabolic processes occur in parallel, proteins and particles jiggle around wildly. If, however, those cells do not get enough nutrients, their energy level drops. This leads to a marked decrease of the cytoplasmic pH -- the cells acidify. In response, cells enter into a kind of standby mode, which enables them to survive. A team of researchers from Dresden, Germany, have found out that the cytoplasm of these seemingly dead cells changes its consistency from liquid to solid. Thereby, they protect the sensitive structures in the cellular interior.
Cells can enter into a kind of standby mode -- called dormancy -- when confronted with unfavorable conditions such as nutrient deprivation. In this state, cells drastically reduce their metabolism and shut down growth and cell division. In extreme cases, such cells are hardly or not at all distinguishable from dead cells -- and yet they can re-emerge from this state unharmed and continue to grow and divide when conditions in their environment improve.
Munder and colleagues from Dresden (Germany) under supervision of Simon Alberti wanted to understand how cells switch on and off the standby mode. They focused their efforts on yeast cells, which they observed during starvation. Their observation: The cytoplasm loses its dynamics, cell organelles and particles slow down and many proteins form large, microscopically visible structures. It seems as if the cytoplasm changes its consistency in response to nutrient deprivation. And indeed: a closer look with highly sensitive biophysical methods shows that the material state of the cytoplasm changes from liquid to solid -- the cell enters into a kind of rigor mortis. As it turns out, the cytoplasmic pH, which decreases markedly under starvation conditions, plays a crucial role in this process.
Remarkably, the sleeping cells -- in contrast to dead cells -- can also reverse this process. When nutrients are added back, the pH rises again, the cytoplasm fluidizes and cells continue to grow and divide. The studies of Munder and colleagues show that the state of the cytoplasm is crucial for switching on and off the standby mode: ''Cells seem to have a control mechanism in place, which they use for the regulation of their material properties in response to certain environmental cues, thereby ensuring their survival''. Thus, it seems to be possible to trick death by shutting down all processes of life in a controlled manner. Whether this trick can be taught to human cells will become clear in the next couple of years.
Story Source:
The above story is based on materials provided by Max-Planck-Gesellschaft. Note: Materials may be edited for content and length.
Journal Reference:
Matthias Christoph Munder, Daniel Midtvedt, Titus Franzmann, Elisabeth Nüske, Oliver Otto, Maik Herbig, Elke Ulbricht, Paul Müller, Anna Taubenberger, Shovamayee Maharana, Liliana Malinovska, Doris Richter, Jochen Guck, Vasily Zaburdaev, Simon Alberti. A pH-driven transition of the cytoplasm from a fluid- to a solid-like state promotes entry into dormancy.eLife, 2016; 5 DOI: 10.7554/eLife.09347
Source : ScienceDaily
Friday, 25 March 2016
Download Pharmaceutical Biotechnology Book
Pharmaceutical Biotechnology
Identifier PharmaceuticalBiotechnology
Identifier-accesshttp://archive.org/details/PharmaceuticalBiotechnology
Identifier-ark ark:/13960/t14n09v94
Ppi 300
Ocr ABBYY FineReader 8.0
Language English
Collection opensource
comment
Monday, 14 March 2016
Hydra can modify its genetic program

- Y. Wenger, W. Buzgariu, B. Galliot. Loss of neurogenesis in Hydra leads to compensatory regulation of neurogenic and neurotransmission genes in epithelial cells. Philosophical Transactions of the Royal Society, November 2015 DOI:10.1098/rstb.2015.0040